

01 Introduction to the study
Eukaryotic cells pack up to 2 meters of genomic DNA up to 2 meters in a nucleus with a diameter of a few microns through a fractionated DNA-protein assembly compression. The first stage is the nucleosome, which consists of 147 bp of DNA wrapped in an octamer of four core histones (H2A, H2B, H3, and H4). This basic repeating unit of the nucleosome is then organized into 10-nanometer "beaded" chromatin fibers that are further compressed into higher-order chromatin structures to accommodate micron-sized nuclei. The organization of chromatin is regulated by a large number of chemical modifications, especially in the N-terminal tail of histone core proteins, such as acetylation and methylation. Histone modifications regulate the packaging of nucleosomes into higher-order chromatin structures to affect genomic DNA accessibility to transcriptional mechanism proteins. Subsequently, chromatin compression in different epigenomic states controls their gene expression and has a significant impact on many cellular processes, such as DNA replication, cell division, DNA damage, and DNA repair.
How different histone modifications shape higher-order chromatin structures in each epigenomic state remains an important question. Due to the limited resolution of conventional light microscopy, current understanding of higher-order chromatin structures defined by different histone modifications is indirectly inferred by in vitro biochemical analysis, such as chromatin immunoprecipitation (ChIP). These analyses often rely on the analysis of fragments of DNA from aggregated cell populations and lose information at the single-cell level. Currently, recent advances in super-resolution fluorescence microscopy have made it possible to image chromatin structures with sub-diffraction finite resolution in fixed and living cells. Location-based super-resolution microscopy such as (direct) stochastic optical reconstruction microscopy (STORM) provides an optimal spatial resolution to directly visualize previously invisible higher-order chromatin structures or even single nuclei in vivo with an optical resolution of 20-30 nm. Super-resolution imaging shows that in vivo chromatin structures consist of heterogeneous nucleosome clusters, as well as different chromatin packages for different epigenomic states at specific gene loci. However, in situ genome-wide higher-order chromatin structures formed by different histone modifications remain elusive.
The study focuses on the comprehensive in situ characterization of genome-wide higher-order chromatin structures defined by histone acetylation and methylation markers, as well as their spatial proximity, to co-form the chromatin environment of a single mammalian nucleus through STORM. We selected a set of 10 histone labels, including lysine acetylation involved in active transcription and lysine methylation involved in inhibition and active transcription. Super-resolution imaging and quantitative analysis of the study revealed three main structural features of higher-order chromatin: histone acetylation to form spatially separated nucleosome nanoclusters, active histone methylation to form spatially dispersed nucleosome nanodomains, and inhibitory histone methylation to form highly concentrated large aggregates. Bicolor STORM imaging showed transcriptionally active histone labeling consistent with "open" chromatin and transcriptional inhibition histone labeling consistent with highly concentrated chromatin. Further studies of their spatial proximity show that inhibitory and active histone markers mostly have spatial exclusivity, while considerable colocalization can be observed in active histone labeling. In summary, super-resolution imaging helps to reveal how histone acetylation and methylation form higher-order chromatin structures at the level of individual mammalian nuclei at scales ranging from tens of nanometers to a few microns.
02Research Results (Excerpt)
1. STORM images and quantitative characterization of different histone labels
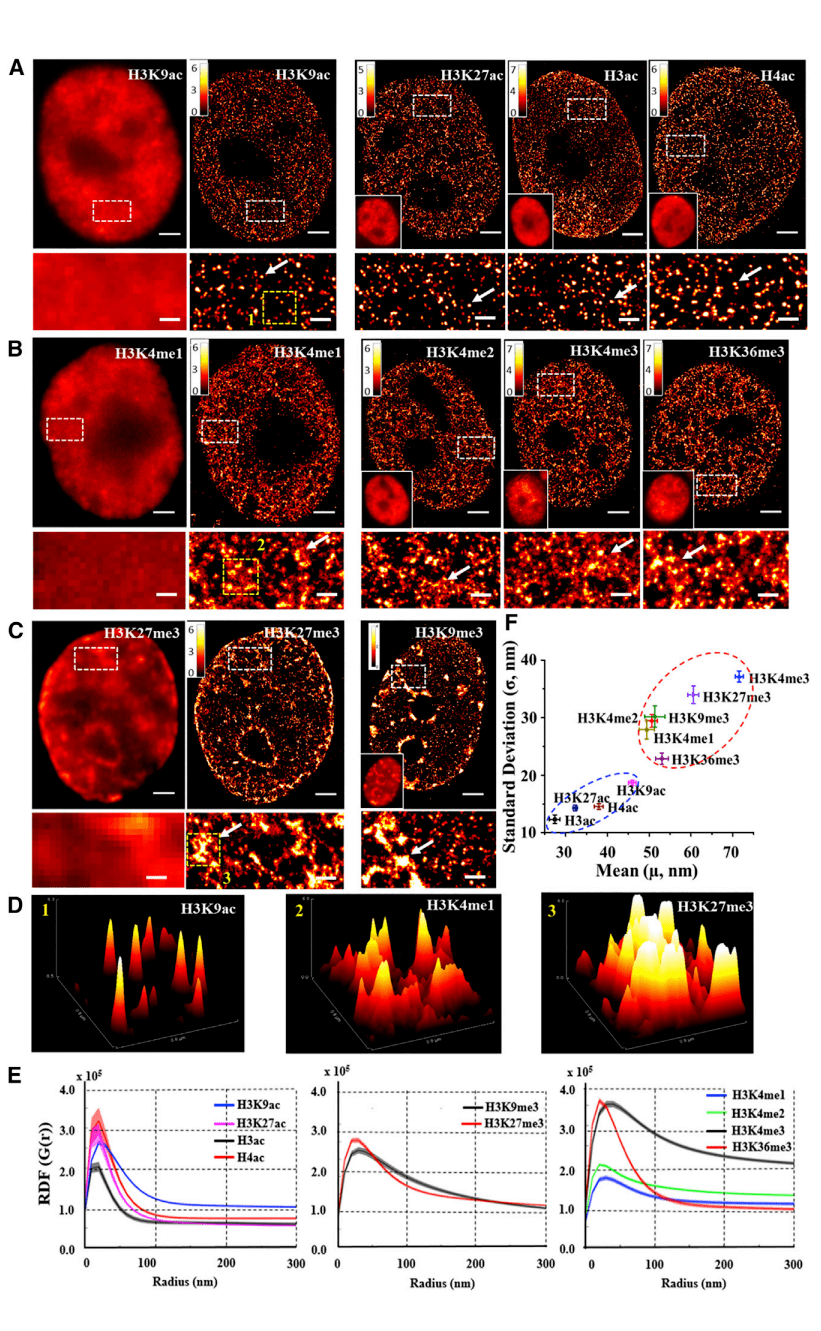
The authors visualized genome-wide higher-order chromatin structures defined by 10 histone modifications in mammalian nuclei. Figures 1A-1C show representative widefield and super-resolution images from 10 histone labels, including transcriptionally active histone acetylation markers (H3K9ac, H3K27ac, H3ac, and H4ac), transcriptionally active histone methylation markers (H3K4me1, H3K4me2, H3K4me3, and H3K36me3), and transcriptional inhibitory histone methylation markers (H3K27me3 and H3K9me3). The super-resolution images of Figures 1A and 1B show obvious structural features: histone acetylation labeling forms spatially separated and discrete nucleosome nanoclusters, and histone methylation labeling forms highly heterogeneous and spatially dispersed nucleosome nanodomains. The wide-field image of Figure 1C shows inhibitory markers (H3K27me3 and H3K9me3) as concentrated aggregates within the nucleus. Super-resolution images clearly show the presence of even very large (micron-sized) nuclei in the nuclei, nucleoli, and the periphery of the nucleus. Figure 1D shows the overall distribution of three representative histone markers (H3K9ac, H3K4me1, and H3K27me3) in the selected super-resolution image region in Figures 1A-1C, which clearly shows three distinct features of higher-order chromatin structures.
The authors quantified these structural features formed by each histone marker. Figure 1E shows that histone acetylation markers exhibit narrow spikes on short length scales of less than 50 nm, indicating the presence of highly aggregated small nanostructures; The RDF distribution of histone methylation markers is wider, indicating the presence of larger polymers and longer associated lengths. Further quantifying the size of nanoclusters and nanodomains formed by different histone labels, Figure 1F shows a scatterplot of average size vs. SD.
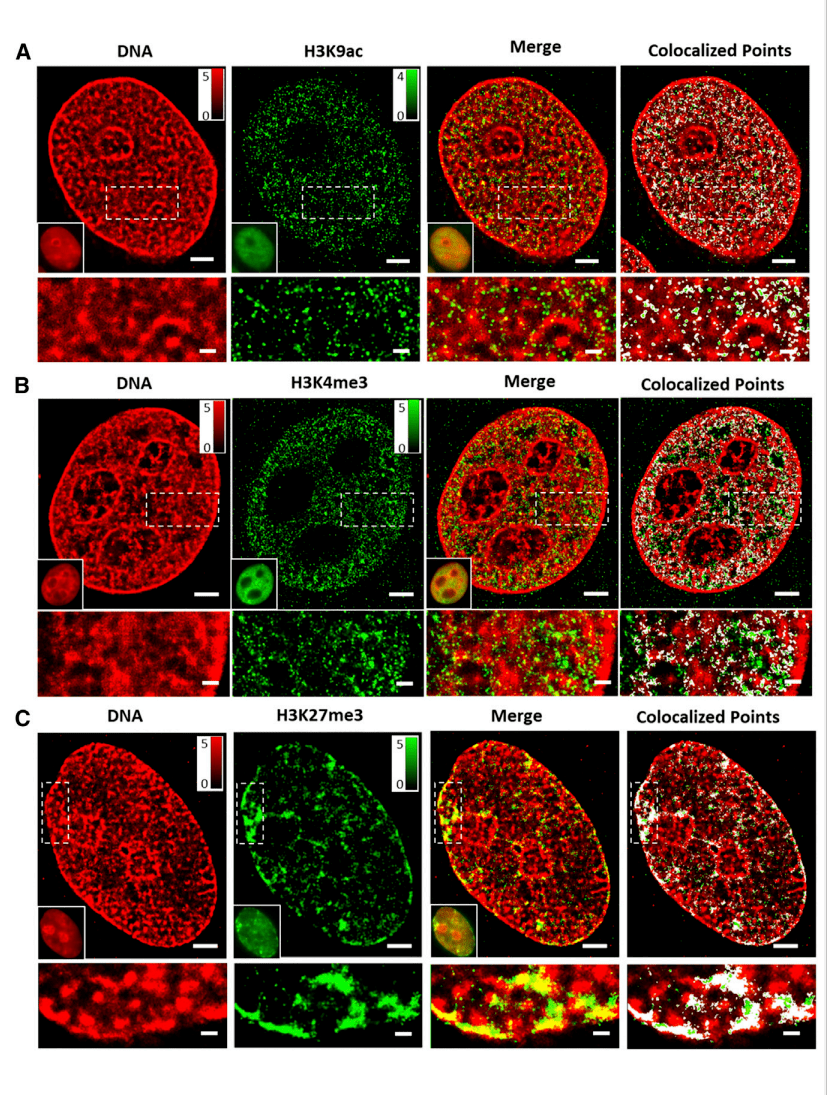
2. Two-color STORM image of different histone markers and DNA in the nucleus of the interphase cell
The authors observed the spatial relationship between histone markers and DNA using two-color STORM imaging. Figure 2 shows representative super-resolution images of transcriptionally active or inhibitory histone markers (green, labeled with Cy3B) and DNA (red, labeled with Alexa Fluor 647), along with their merged images and colocators (labeled with white). These images show that DNA forms separated regions in a highly compact manner in the nucleus, where active histone markers for H3K9ac or H3K4me3 are more abundant in less dense regions of DNA (Figures 2A and 2B), while inhibitory histone labeling for H3K27me3 mostly coincides with concentrated regions of DNA (Figure 2C). This direct visual evidence shows the relationship between histone labeling and chromatin density: inhibitory histone labeling is associated with highly concentrated chromatin structures, while active histone labeling is associated with more "open" or less dense chromatin structures.
03 Research Summary
The authors demonstrate the potential of the super-resolution positioning microscope STORM to directly visualize genome-wide higher-order chromatin structures in epigenomic states in interphase mammalian nuclei. The results reveal previously unseen unique features of higher-order chromatin structures formed by histone acetylation and methylation markers. This result lays the groundwork for future studies of the functional significance of these structural features and how they change in different disease states.
04 Super-resolution microscopic imaging system iSTORM
The STORM imaging technology mentioned above has been successfully commercialized, and there are experts and teachers who need STORM imaging technology for experimental research, please communicate and make an appointment
The super-resolution microscopy imaging system iSTORM has successfully achieved a breakthrough in the diffraction limit of optical microscopy, making it possible to engage in single-molecule localization and counting of biological macromolecules, subcellular and supramolecular structure analysis, and biodynamics of biological macromolecules on a resolution scale of 20 nm, thus bringing major breakthroughs to life sciences, medicine and other fields.

Ultra-high resolution microscopy imaging system iSTORM has the characteristics of 20 nm ultra-high resolution, 3-channel simultaneous imaging, 3D simultaneous shooting, real-time reconstruction, and 2-hour novice mastery, etc., has realized the localization and counting of single molecules in living cells, and provides fluorescent dye selection, sample preparation, imaging services and experimental solutions as an overall solution. It has been highly recognized by more than 50 scientific research groups and more than 100 researchers.
References:
Xu J , Ma H , Jin J , et al. Super-Resolution Imaging of Higher-Order Chromatin Structures at Different Epigenomic States in Single Mammalian Cells[J]. Cell Reports, 2018, 24(4):873.







